Who hasn’t played a game of “Guess Who?” as a child? Enrico Borriello and Bryan Daniels, faculty in ASU’s new School of Complex Adaptive Systems, argue that scientists still do, in the form of gene reprogramming experiments.
The goal of “Guess Who?” is to identify a hidden character by asking the fewest number of yes/no questions: I might ask if your character wears a hat, and if you say yes, remove all the characters not wearing one. The goal of gene reprogramming, on the other hand, is to change cell types from one to another. What do these two problems have in common?
Inside cells, a complex dance of proteins creates distinct patterns that lead to different cell types. For example, heart cells make a different set of proteins than liver cells, while relying on the same genetic code. In a cell, we might aim for a certain cell type by forcing the presence of proteins that are specific to that type of cell.
In a game of “Guess Who?”, I can guarantee that the character I identify will wear a hat by simply restricting my search to those who are wearing one. Similarly, if I want a cell type that has a particular set of proteins, I might start by simply forcing the cell to make those proteins.
How far can we take this analogy? In biology, the problem is complicated by the fact that cells are known to react to changes in complicated ways. Our initial attempts at control could fundamentally alter the cell’s behavior, creating new sets of proteins that were never previously produced together. A “Guess Who?” character wearing a hat can’t avoid detection by recruiting new hat-wearing characters after the question is asked—but cells may change after forcing.
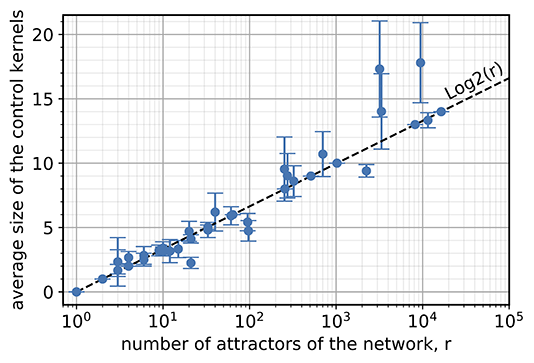
Unexpectedly, the researchers found that genetic networks reacted in a way that did not require much additional control. Studying a database of known systems in cell biology, they calculated the number of components that would need to be controlled to guarantee particular final states. This included systems that generate cell types, cell death, cell cycles, cell signaling, and the effects of cancer. Across all 49 of the examples tested, they found a similar result—a hypothetical controller would only need to force the behavior of a few components to get a desired collective behavior.
Still, the question remained of why this easy control was so pervasive. The researchers found that the size of the genome is irrelevant. Instead, the number of proteins that need to be controlled increases very slowly only when the number of possible cell types increases. This is just like “Guess Who?”—if at each step, I can eliminate a large fraction of the remaining characters, then the number of questions I need to win is always relatively small (more specifically, the logarithm of the number of characters).
As a final twist, the researchers were surprised to find that this seemingly simple problem has been puzzling mathematicians for years. A peculiar example called the “projective plane” can create sets of characters that require many more questions to specify than one would naively expect (see example diagram below). These examples are related to the idea of a Latin square, which is similar to a Sudoku puzzle. These strange cases are rare, but their exact number is still unknown.
Importantly, the abstract features that lead to easy control are the same for all types of network dynamics, not just those describing genetic regulation. For this reason, the researchers anticipate that future applications could appear not just in biology but across other complex systems studied in the School, including social and engineered systems.
For more information, see Borriello and Daniels’ recent publication in Nature Communications (https://doi.org/10.1038/s41467-021-25533-3).